Many teams run into the same problem: a knockout gives you a phenotype, but not the answer. Most modern research questions require understanding what one specific variant does—not what happens when the entire gene is removed. That’s why CRISPR knock-in has become essential: it’s the only editing approach capable of testing precise, mechanistic hypotheses without disrupting the whole system.
This article focuses on two things researchers care about most:
- When knock-in is the only way to get a reliable, biologically meaningful result, and
- How to avoid the common traps that make knock-in slow, inconsistent, or frustrating.
If your work involves SNPs, signaling dynamics, drug-response pathways, or allele-specific effects, these two questions directly determine whether your experiments succeed.
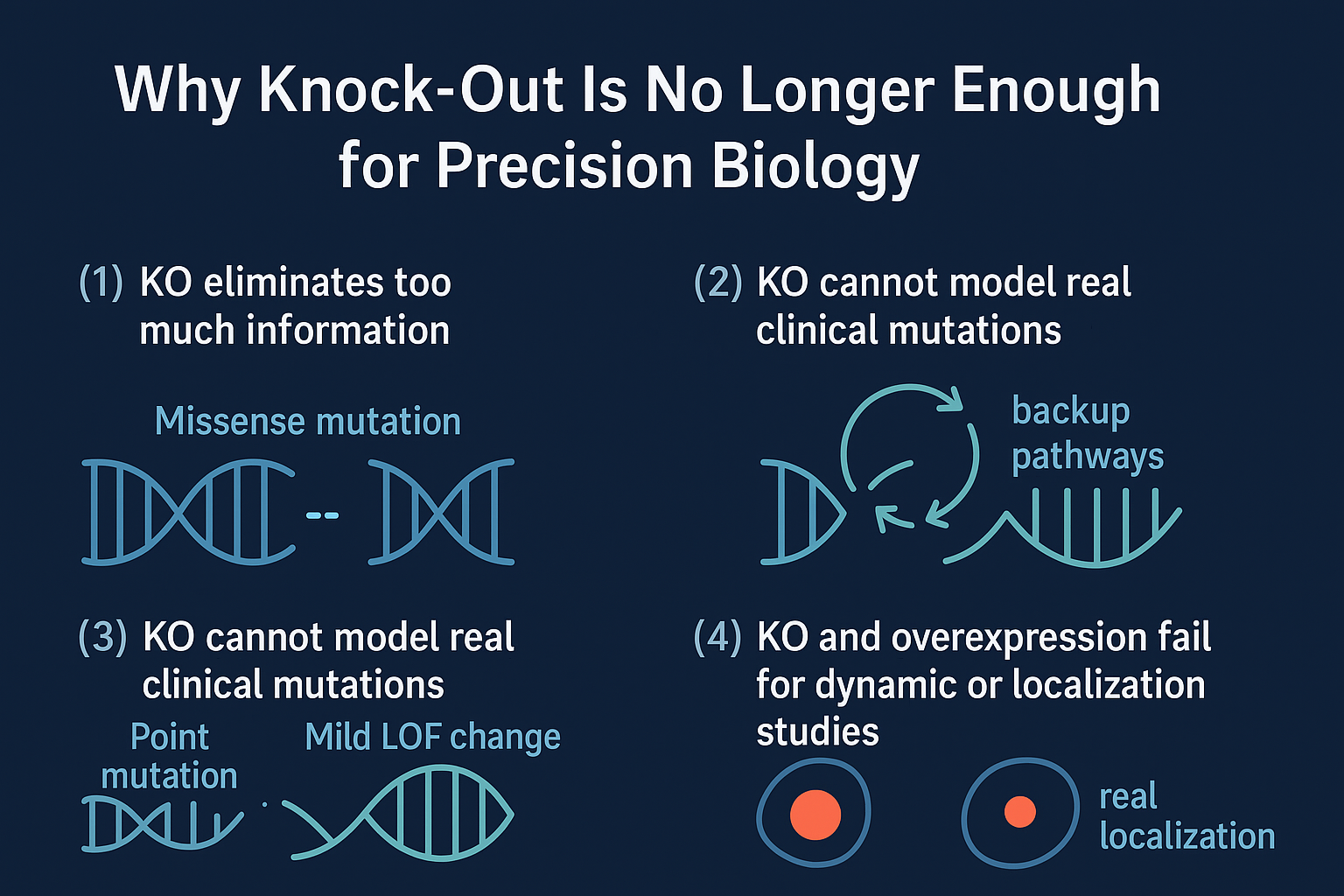
Why Knock-Out Is No Longer Enough — and Why CRISPR Knock-In Matters
Knock-out is excellent at answering one type of question:
“Is the gene essential?”
But most modern projects focus on “What does this variant actually do?”
—and KO simply cannot answer that.
Here’s what researchers constantly run into:
KO eliminates too much information
A missense mutation might weaken a domain slightly. KO erases the domain, the protein, and its entire regulatory context.
This is why KO phenotypes often look dramatic while patient mutations do not.
KO frequently triggers compensatory pathways
Delete a gene in a signaling pathway, and 24–48 hours later, backup pathways light up.
What you measure is the cell responding to stress—not the variant you care about.
KO cannot model real clinical mutations
Most clinically relevant variants are:
- point mutations
- mild LOF changes
- splice alterations
- subtle domain disruptions
And this is exactly where precise knock-in editing becomes essential.
To introduce defined mutations at the endogenous locus—without disrupting the surrounding regulatory context—many teams now rely on CRISPR knock-in workflows.
If you want to explore how these precise edits are typically generated, you can click here to see how CRISPR knock-in is applied in real research settings.
KO and overexpression fail for dynamic or localization studies
Reviewers often push back on overexpression imaging:
“Is this localization real, or an artifact?”
Endogenous context matters—and KO/OE systems don’t provide it.

What CRISPR Knock-In Enables That Knock-Out Cannot — and Why Validated Knock-In Cell Lines Are Becoming Essential
A simple rule captures the difference:
Knock-in alters exactly the part you want to study.
Knock-out destroys the entire neighborhood.
Here’s why knock-in is now essential in mechanism-level biology:
Modeling true patient mutations
Drug sensitivity, resistance, and pathway rewiring often hinge on a single codon.
Knock-in is the only accurate way to model these subtle effects.
Endogenous tagging with real biological timing
Imaging and localization experiments become dramatically cleaner.
You no longer question whether a signal is biological or an artifact.
Stable, drift-free pathway reporters
Endogenous reporters avoid the transcriptional drift common in plasmid-based systems after 24–48 hours.
Precise structure–function interrogation
Phospho-mimics, domain disruptions, and binding-site tests all rely on precise, allele-level edits.
Allele-specific analysis
KO eliminates both alleles; knock-in lets you modify one and leave the other untouched—critical for imprinting and SNP-driven effects.
As these applications expand, many groups no longer want to spend 6–12 weeks troubleshooting HDR or screening mosaic clones. Instead, they increasingly turn to validated knock-in cell lines that already contain defined point mutations or regulatory edits. To see how these models are built and applied in variant-level research, you can click here for examples of well-characterized knock-in cell lines used in pathway and disease-mutation studies.
The Increasing Demand for Reliable Knock-In Cell Lines
Ask any PI who runs multiple projects:
“Knock-in is powerful, but we can’t risk the entire quarter on one HDR attempt.”
And the concerns are justified.
Timelines are longer—and more fragile—than most planning charts assume
Even in friendly lines like HEK293, the process commonly requires:
- 1–2 weeks for design
- 2–4 weeks for editing + recovery
- 2–4 weeks for clonal screening and validation
A single mosaic clone can double the timeline.
HDR success is highly cell-line dependent
The same sgRNA can show ~20–40% HDR in one cell line and <5% in another.
iPSCs are especially tricky.
Validated lines give reproducibility KO/OE models cannot match
This is why KI models become long-term assets: they support drug testing, mechanistic studies, imaging, and variant comparisons.
Complex biological questions simply demand knock-in
Drug-resistance variants, regulatory element testing, and pathway sensors cannot be approximated by KO or OE approaches.
Practical Advantages That Matter to Every Lab
Researchers don’t adopt knock-in because it’s elegant—they adopt it because it solves real problems.
Better experimental clarity
The data becomes easier to interpret because the context is endogenous.
Stronger reproducibility across projects
Stable KI models hold up across students, batches, and assays.
Expanded experimental toolbox
Only KI supports:
- precise pathway reporters
- allele-specific measurements
- phospho-mimic modeling
- live-cell tracing under physiological regulation
Lower long-term cost
A single good KI line may support five or more projects.
Shorter timelines when using optimized methods or validated models
Donor refinement, HDR-enhancing strategies, and well-designed screening pipelines drastically reduce trial-and-error.
What Researchers Should Consider When Planning a Knock-In Strategy
To avoid wasted cycles and failed clones, researchers should have these components clear before starting:
What exact edit do you need?
Point mutation? Tag insertion? Exon replacement?
These dictate donor type, arm length, and validation strategy.
Is your cell line HDR-friendly?
Some lines require synchronization; others demand electroporation.
Immortalized lines are usually easier than iPSCs.
Donor template design determines success
Silent mutations, cut-blocking edits, and homology arm symmetry matter more than many expect.
Validation must be planned early
NGS for PMs, protein-level validation for tags, PCR + sequencing for exon swaps.
Assess timeline, risk, and opportunity cost
When projects are time-sensitive or mechanistically central, validated KI lines dramatically reduce risk.
Conclusion — Knock-In Is Becoming the Standard for Precision Biology
As questions in molecular biology become more precise, knock-in is shifting from a “specialized technique” to a default requirement for mechanistic studies. It offers cleaner insights, more stable models, and biologically faithful results that knock-out and overexpression strategies cannot match.
If your project involves variant effects, signaling dynamics, patient-derived mutations, or pathway modeling, knock-in isn’t optional—it’s essential.
The future of gene research won’t be defined by whether we can delete a gene, but by something more fundamental:
Whether we can make the exact change biology requires.

